Importing HDF5 Files
HDF5 (Hierarchical Data Format version 5) is a file format designed to store and organize large and complex datasets like images, time series, etc. An HDF5 file serves as a container that holds a variety of data objects, including groups, datasets, and datatypes. Its tree-like structure helps organize multiple data objects efficiently, with support for compression and encryption.
DolphinDB has developed an HDF5 plugin to import HDF5 datasets into DolphinDB and supports data type conversions. This page explains how to use the plugin to import HDF5 datasets into DolphinDB.
Installing HDF5 Plugin
The HDF5 plugin is pre-installed by default and can be loaded simply by running the following script.
loadPlugin("hdf5")HDF5 Data Import Methods
The DolphinDB HDF5 plugin provides the following methods to facilitate HDF5 file import:
loadHDF5: Imports a specified HDF5 dataset as an in-memory table.loadPandasHDF5: Imports specified HDF5 dataset, saved by pandas, as an in-memory table.loadHDF5Ex: Imports HDF5 datasets directly into either a distributed database or in-memory database.HDF5DS: Divides a HDF5 dataset into multiple smaller data sources. It can be used with functionmrto load data.
The following table outlines common parameters of these methods.
| Parameter | Description |
|---|---|
| filename | A string indicating the directory of input HDF5 file. |
| datasetName | A string indicating the dataset name, i.e., the table name. |
| schema |
A table with the following columns:
|
| startRow | An integer indicating the start row from which to read the HDF5 dataset. If not specified, the dataset will be read from the beginning. |
| rowNum | An integer indicating the number of rows to read from the HDF5 dataset. If not specified, the reading continues until the end of the dataset. |
Getting Started
Loading to Memory and Importing to Database
Like loadText, loadHDF5 imports data as
in-memory tables, but requires you to specify which dataset to import from the
HDF5 file.
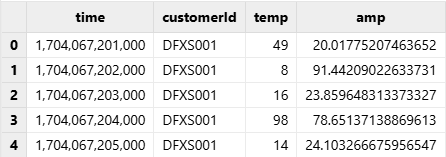
The following example demonstrates how to import the “data” dataset from demo1.h5.
dataFilePath = "./hdf5file/demo1.h5"
tmpTB = hdf5::loadHDF5(dataFilePath, "data");
select * from tmpTBOutput:
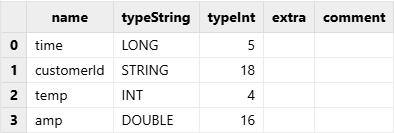
Call the schema function to view the table schema (column names,
data types, etc):
tmpTB.schema().colDefs;Output:
For HDF5 files saved by pandas, use loadPandasHDF5.
To import data to database, use tableInsert or
append! functions. Ensure that the target database and
table are already created before proceeding.
Script for creating the database and table:
create database "dfs://demoDB" partitioned by HASH([SYMBOL,25])
create table "dfs://demoDB"."data"(
time LONG,
customerId SYMBOL,
temp INT,
amp DOUBLE
)
partitioned by customerIdIn the original data, the time column is of type LONG. To demonstrate how to import data into the database, the time column has been set to LONG during the database and table creation. Detailed instructions on converting the temporal type can be found in the section below.
Then insert the table to the database:
dfsTable = loadTable("dfs://demoDB", "data")
tableInsert(dfsTable, tmpTB)Importing to Database Directly
In addition to explicitly using in-memory tables for database import, DolphinDB
provides loadHDF5Ex function that combines loading, cleaning,
and storage into one streamlined process. Ensure that the target database and
table are already created before proceeding.
The following example demonstrates how to import demo1.h5 to table “data” in database “demoDB”. To
start with an empty database, use the truncate function to
remove existing rows.
// remove existing rows
truncate("dfs://demoDB","data")
// import to database
dataFilePath = "./hdf5file/demo1.h5"
hdf5::loadHDF5Ex(database("dfs://demoDB"), "data",
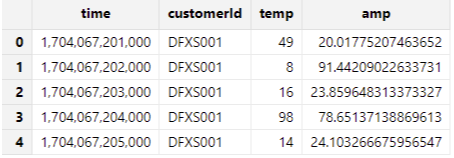
"customerId", dataFilePath, "data");Check the results:
dfsTable = loadTable("dfs://demoDB","data")
select * from dfsTable Output:
Unlike loadHDF5, loadHDF5Ex requires users to
specify database details including database name, table name, and partitioning
columns. Besides, loadHDF5Ex offers transform parameter
to clean or process data directly during database import. For more informatin
about transform parameter, see Data Cleaning and Preprocessing.
Handling Column Names and Data Types
The HDF5 plugin automatically retrieves data types from file attributes during import and converts them to the corresponding DolphinDB data types. Details of supported HDF5 data types and their conversion rules can be found in Plugins > HDF5 > Data Type Mappings.
While automatic detection is generally reliable, occasional manual adjustments by modifying schemaparameter may be needed.
Extracting HDF5 File Schema
Users can use the extractHDF5Schema function to preview a HDF5
file's schema before data loading, including details like column names, data
types, and the number of columns. It helps users identify potential issues such
as mismatched data types and modify the schema accordingly for proper data
loading.
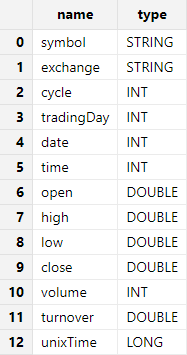
For example, use extractHDF5Schema to get the schema of the
sample file hdf5Sample.h5:
dataFilePath = "./hdf5file/hdf5Sample.h5"
hdf5::extractHDF5Schema(dataFilePath,"sample")Output:
Specifying Column Names and Types
If the column names or data types automatically inferred by the system are not
expected, we can specify the column names and data types by modifying the schema
table generated by function extractHDF5Schema or creating the
schema table directly.
Specifying Column Names
For example, use the following script to modify schema and rename column “symbol” as “securityID” :
dataFilePath = "./hdf5file/hdf5Sample.h5"
schemaTB = hdf5::extractHDF5Schema(dataFilePath,"sample")
update schemaTB set name = ["securityID","exchange","cycle","tradingDay","date","time","open","high","low","close","volume","turnover","unixTime"]Use function loadHDF5 to import an HDF5 file and specify
schema parameter as the modified schemaTB.
tmpTB = hdf5::loadHDF5(dataFilePath, "sample", schemaTB);Call function schema to view the table schema. Column “symbol”
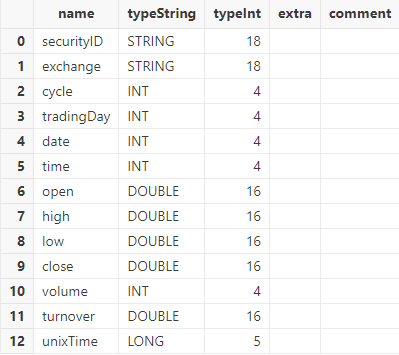
has been renamed as “securityID”.
tmpTB.schema().colDefs;Output:
Specifying Data Type
The time columns in hdf5Sample.h5 are stored as timestamps of type INT. However, the expected data type for column tradingDay and date is DATE, and for unixTime, it is TIMESTAMP. To convert the the data type of time columns:
dataFilePath = "./hdf5file/hdf5Sample.h5"
schemaTB = hdf5::extractHDF5Schema(dataFilePath,"sample")
update schemaTB set type="DATE" where name="tradingDay" or name="date"
update schemaTB set type="TIMESTAMP" where name="unixTime"Use function loadHDF5 to import the HDF5 file and specify
schema parameter as schemaTB.
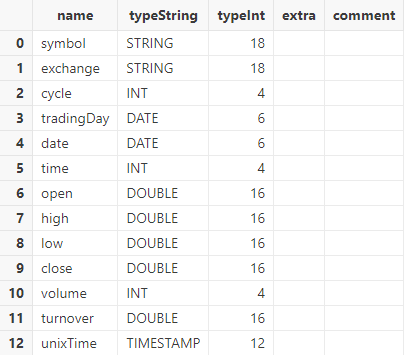
tmpTB = hdf5::loadHDF5(dataFilePath, "sample", schemaTB);Call the schema function to view the table schema:
tmpTB.schema().colDefs;The data type of column tradingDay and date has been converted to DATE, and unixTime to TIMESTAMP.
Importing Specific Rows
loadHDF5 method supports importing specific rows by specifying
startRow and rowNum parameters.
To import 100 rows starting from row 10:
tmpTB = hdf5::loadHDF5(dataFilePath, "sample", schemaTB, 9, 100);Check the total rows:
select count(*) from tmpTB
/*
count
-----
100
*/Check the first 5 rows:
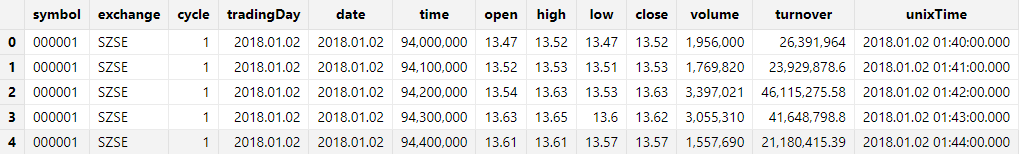
select top 5 * from tmpTB;Output:
Data Cleaning and Preprocessing
DolphinDB provides various built-in functions for common data cleaning task, including handling missing values, duplicate values, and outliers, and performing data normalization and standardization. This section focuses on several common scenarios.
HDF5 plugin provides two methods for importing data: loadHDF5
function and loadHDF5Ex function (See Getting Started). While both methods support
basic data cleaning, loadHDF5Ex introduces a distinctive
transform parameter that enables data cleaning and preprocessing during
data import. This parameter accepts a unary function that processes the loaded table
and returns a cleaned dataset, which is then directly written to the distributed
database without intermediate memory storage. Our focus will be on practical
applications of the transform parameter in loadHDF5Ex.
Data Type Conversion of Temporal Types
Time columns are typically stored as type INT or LONG in HDF5 files. Directly
loading time columns of type INT will lead to incorrect results. For example,
9:31:00.000 is stored as 93100000 in hdf5Sample.h5. In such case, one way is to use
the transform parameter of the loadHDF5Ex function to
wrap the replaceColumn! function for data type conversion.
First, create a distributed database and a table.
dataFilePath = "./hdf5file/hdf5Sample.h5"
dbPath = "dfs://testDB"
db = database(dbPath,VALUE,2018.01.02..2018.01.30)
schemaTB = hdf5::extractHDF5Schema(dataFilePath,"sample")
update schemaTB set type="DATE" where name="tradingDay" or name="date"
update schemaTB set type="TIME" where name="time"
update schemaTB set type="TIMESTAMP" where name="unixTime"
tb = table(1:0,schemaTB.name,schemaTB.type)
pt = db.createPartitionedTable(tb,`tb1,`date);Define a user-defined function i2t to encapsulate the data
processing logic, converting the time column to TIME type.
def i2t(mutable t){
replaceColumn!(table=t, colName=`time, newCol=t.time.format("000000000").string().temporalParse("HHmmssSSS"))
return t
}When processing data in a user-defined function, please try to use in-place modifications (functions that finish with !) if possible for optimal performance.
Call function loadHDF5Ex and assign function
i2t to parameter transform. The system executes
function i2t with the imported data, and then saves the result
to the database.
schemaTB = hdf5::extractHDF5Schema(dataFilePath,"sample")
update schemaTB set type="DATE" where name="tradingDay" or name="date"
update schemaTB set type="TIMESTAMP" where name="unixTime"
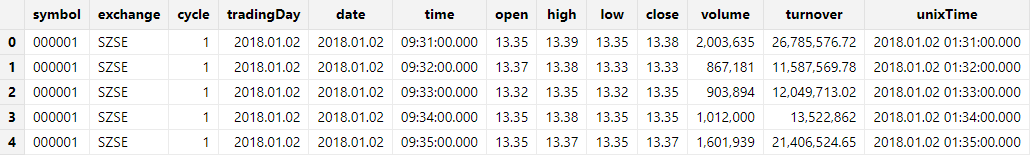
hdf5::loadHDF5Ex(database("dfs://testDB"), "tb1", "date", dataFilePath,"sample", schemaTB, , , i2t)Check the results:
select top 5 * from loadTable("dfs://testDB", "tb1")Column 'time' is stored as data type TIME instead of INT.
Handling Null Values
Efficient handling of missing values is essential when working with real-world
datasets. DolphinDB offers built-in functions like bfill,
ffill, interpolate, and
nullFill, enabling quick and effective handling of
incomplete data in tables. As parameter transform only takes a function
with one parameter, to assign a DolphinDB built-in function with multiple
parameters to transform, we can use partial
application to convert the function into a function with only one
parameter. To fill null values xxx, the following statement can be passed as
parameter transform:
nullFill!(t,0)Importing Multiple HDF5 Files or Datasets in Parallel
An HDF5 file may have multiple datasets. Therefore, DolphinDB supports parallel import of both HDF5 files and datasets. While multi-file import is similar to text files, we'll focus on parallel dataset import.
First, use lsTable to list all datasets in the file.
dataFilePath = "./hdf5file/demo1.h5"
datasets = hdf5::lsTable(dataFilePath).tableNameDefine a data import function and use the submitJob function to
assign a thread for each dataset, enabling background batch data import:
def writeData(dbName,tbName,file,datasetName){ //write data to database
t = hdf5::loadHDF5(file,datasetName)
replaceColumn!(table = t, colName = `time, newCol = timestamp(t.time)) //convert data type
tableInsert(loadTable(dbName,tbName), t)
}
dbName = "dfs://demoDB_multi"
tbName = "data"
for(datasetName in datasets){
jobName = "loadHDF5" + datasetName
submitJob(jobName, jobName, writeData{dbName, tbName}, dataFilePath, datasetName)
}Use the select statement to calculate the time taken for parallel
batch datasets import:
select max(endTime) - min(startTime) from getRecentJobs(2);Importing a Single Large HDF5 File
To avoid memory overflow (OOM) when importing large files, use the
HDF5DS function to split the file into smaller chunks for
import. Unlike textChunkDS, HDF5DS allows setting
the exact number of data sources to evenly divide the dataset.
First, split the sample data into two equal parts by setting dsNum to 2.
dataFilePath = "./hdf5file/demo1.h5"
datasetName = "data"
ds = hdf5::HDF5DS(dataFilePath, datasetName, , 2)Then, use the mr function to write the data into the database:
truncate(dbUrl = "dfs://demoDB",tableName = "data")
pt = loadTable("dfs://demoDB", "data")
mr(ds = ds, mapFunc = append!{pt}, parallel = false)Importing Datasets of Composite Datatypes
DolphinDB supports importing complex HDF5 dataset structures, specifically with:
- Compound type (H5T_COMPOUND)
- Array type (H5T_ARRAY)
- Combination of compound and array types
These composite datatypes require specialized handling during the import process to ensure accurate data representation in DolphinDB.
The following examples will demonstrate the import mechanisms for these complex HDF5 dataset structures.
Compound type
Given an HDF5 dataset contains compound type data:
| 1 | 2 | |
|---|---|---|
| 1 | struct{a:1 b:2 c:3.7} | struct{a:12 b:22 c:32.7} |
| 2 | struct{a:11 b:21 c:31.7} | struct{a:13 b:23 c:33.7} |
When importing a compound type HDF5 dataset, DolphinDB converts the structure into a table where each compound type element is translated into a row. The compound type keys serve as column names, and their values populate the table using DolphinDB's type conversion rules.
The resulting DolphinDB table is structured as follows:
| a | b | c |
|---|---|---|
| 1 | 2 | 3.7 |
| 11 | 21 | 31.7 |
| 12 | 22 | 32.7 |
| 13 | 23 | 33.7 |
Array Type
| 1 | 2 | |
|---|---|---|
| 1 | array(1,2,3) | array(4,5,6) |
| 2 | array(8,9,10) | array(15,16,17) |
When importing an array type HDF5 dataset, DolphinDB converts the structure into a table where each array is translated intoa row. Elements with the same index across arrays are grouped into columns, with column names labeled as array_index.
The resulting DolphinDB table is structured as follows:
| array_1 | array_2 | array_3 |
|---|---|---|
| 1 | 2 | 3 |
| 4 | 5 | 6 |
| 8 | 9 | 10 |
| 15 | 16 | 17 |
Combination of Compound and Array Types
Given an HDF5 dataset contains complex combinations of datatypes:
| 1 | 2 | |
|---|---|---|
| 1 |
struct{a:array(1,2,3) b:2 c:struct{d:"abc"}} |
struct{a:array(7,8,9) b:5 c:struct{d:"def"}} |
| 2 |
struct{a:array(11,21,31) b:0 c:struct{d:"opq"}} |
struct{a:array(51,52,53) b:24 c:struct{d:"hjk"}} |
For these complex datatype combinations, a prefix convention is used for column naming: Afor arrays and Cfor compound datatypes.
Each nested element is parsed according to its specific type, maintaining consistency with the conventions for arrays and compound datatypes described above.
The resulting DolphinDB table is structured as follows:
| Aa_1 | Aa_2 | Aa_3 | b | Cc_d |
|---|---|---|---|---|
| 1 | 2 | 3 | 2 | abc |
| 7 | 8 | 9 | 5 | def |
| 11 | 21 | 31 | 0 | opq |
| 51 | 52 | 53 | 24 | hjk |
